|
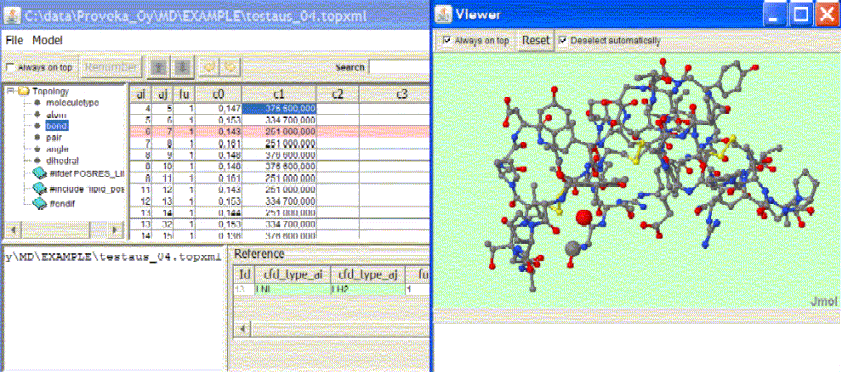
Proveka Molecular Dynamic Proveka Molecular Dynamic is a tool for supporting fast manipulation of molecular data. One of the main challenges in molecular dynamic simulation is to find a suitable set of parametres, especially when developing new molecular structures. Proveka Molecular Dynamic supports this perfectly. User can easily change values and still keep track of the original values. The viewer helps in visualization and locating the atoms. Proveka Molecular Dynamic is GROMACS compatible.
Its main features are techically are
Example 1 : user creates an atom coordinate file and uses PRODRG-server but PRODRG-server changes the atom names. Using Proveka Molecular Dynamic user may in advance make sure that all atoms are named and numbered uniquely. After creating a topology file, user can also check and compare other parametre values from refrerences. Viewer shows exactly which atom, bond, pair or else is under focus. Example 2: user has created a Gromacs coordinate file, a gro-file, with 267724 atoms and a corresponding Gromacs index file, ndx-file. User needs to extract atom numbers that correspond to a criterium to create a few groups for the Gromacs index-file. Example of a gro-file is below: 267724 User enters criteria, for instance POPC and some atom names N, P, C11, etc. Proveka Molecular Dynamic extracts data and generates a partial index-file as follows: 1 2 8 12 13 16 17 20 21 24 53 54 60 64
65 which then can be copied and pasted to the index-file, including a user-defined group name, e.g. [ Group 1 ]. Proveka Molecular Dynamic supports this perfectly! Example 3: user needs to set parametres of an mpd-file. Proveka Molecular Dynamic can be used to create a file and to change the parametre values. Also, it reads an existing mdp-file. Supports default value generation, for instance in energy minimization user sees the defaulf values such as emtol: 10 For more information: Vesa Karhu, CEO, D.Tech. (email first.last(at)proveka.fi) cell phone +358 50 323 4772, Skype: proveka, sales(at)proveka.fi |
Copyright Proveka Oy 2004 -